Last year over 700 boat owners who have non-trailered boats moored in marinas around Aotearoa…
Harnessing passive sampling and DNA extraction innovations for eDNA-based surveillance
A recent study led by University of Otago and Cawthron scientists investigated new techniques for monitoring marine ecosystems using environmental DNA (eDNA). Traditionally, monitoring eDNA requires time-consuming filtration and specialized laboratories, limiting sample replication and in situ species detection (Figure 1). In this study, researchers explored the effectiveness of passive sampling and self-contained DNA extraction methods as alternatives.

Figure 1: Filtration system setup for eDNA sample processing in a lab.
Passive sampling involves using porous materials like artificial sponges and fishing nets to capture eDNA from the water. The results showed that passive sampling yielded comparable fish diversity results to traditional active filtration methods in both controlled and natural marine environments (Fig. 2). This approach eliminates the need for time-consuming vacuum filtration and can be deployed and retrieved quickly by non-scientists.
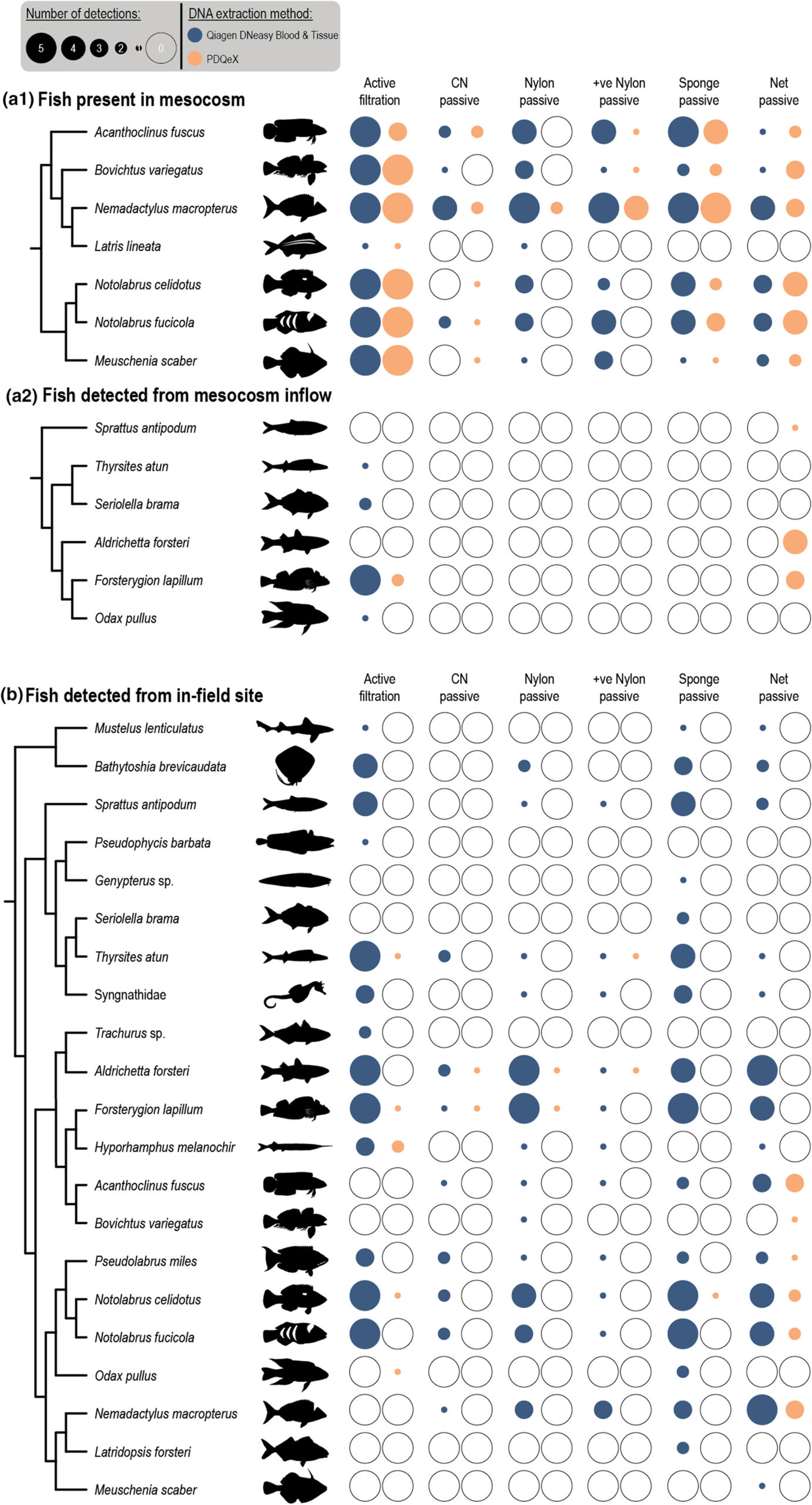
Figure 2: Observed diversity from (a) mesocosm and (b) in situ experiments for each experimental treatment. DNA extraction method is indicated by blue (Qiagen DNeasy Blood & Tissue kit) or orange (PDQeX) circles. Filled and unfilled circles indicate taxon presence or absence. Circle diameter indicates the number of positive detections (maximum 5 replicate for each treatment).
The study also examined the effectiveness of a self-contained DNA extraction system called PDQeX. While the PDQeX system demonstrated promising results in extracting high-abundance DNA in controlled settings, it was less effective for in situ testing in the natural environment. Further optimization is required to improve the system’s ability to detect low-abundance eDNA from the marine environment.
These findings suggest that passive sampling using porous substrates can effectively capture eDNA, simplifying the monitoring process and enabling broader sample replication. However, the choice of the extraction method needs careful consideration, and further refinement of the PDQeX system is necessary for reliable in situ applications.
Overall, this study contributes to the advancement of non-invasive eDNA monitoring methods, offering potential benefits for efficient and scalable assessment of marine ecosystems.




Comments (0)