Marine Biosecurity Toolbox scientist Dr Ulla von Ammon joined the BLAKE Expedition and shared with the expedition team the eDNA tools, including the newly designed Cruising Speed Net, for detecting unwanted organisms in marine environments.
Sampling optimization for molecular biosecurity surveillance – results from an expedition onboard the Robert C. Seamans (SEA, Woods Hole)
Environmental DNA (eDNA) in combination with metabarcoding or real time PCR are promising approaches for marine non-indigenous species (NIS) detection. To maximize detection of rare biodiversity, plankton vertical or horizontal net tows are often used to concentrate eDNA signals over hundreds of liters of volume. The follow-up filtration step of the concentrated sample often becomes a bottleneck in the sample processing pipeline, requiring substantial time and effort. These sample collection and processing steps need to be optimized without affecting the performance of eDNA-based detection for wider end-user uptake and routine implementation for biosecurity surveillance.
In February 2020, our postdoc Ulla von Ammon embarked on the Robert C. Seamans (Picture 1) sailing ship as part of the SEA expedition organized by the Woods Hole Institute to collect seawater samples using different sampling approaches.

The aim of the study was to compare the performance of two sampling approaches (pre-concentration with a Cruising Speed Net versus whole sample collection with a bucket) and different filter types (5 and 1.2 µm GF/C membranes and customized 5 µm self-preserving eDNA filter packs) for targeted species detection and capturing community composition across three New Zealand harbors (Waitemata, Whangarei and Russell; Picture 2).
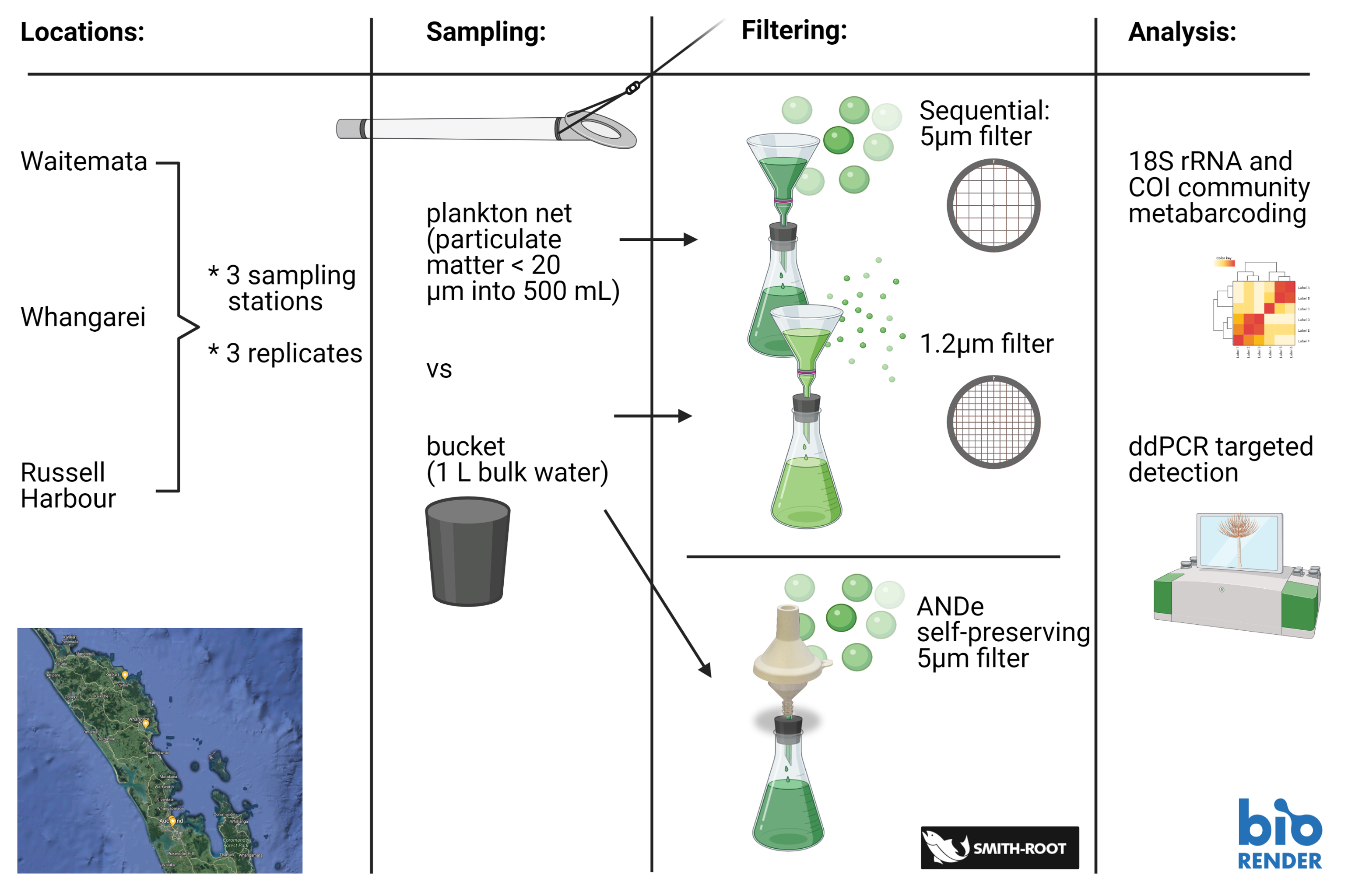
Picture 2: Study design
The metabarcoding data revealed significant differences between the tested sample types, with the bucket sampling combined with 5 µm self-preserving filters better capturing overall species diversity, and macrofauna (marine invertebrates, which includes the main groups for biosecurity interests) better represented in the net samples filtered through 5 µm membranes.
Multispecies detection probabilities were estimated for each workflow using a probabilistic occupancy modelling approach. Highest species richness was attained by the bucket sampling combined with self-preserving filters, comprising a large portion of microplankton. Less diversity but more metazoan taxa were captured in the net samples combined with 5 μm pore size filters. This confirmed the effectiveness of preconcentration plankton for biodiversity screening. The results contribute to streamlining eDNA sampling protocols for uptake and implementation in marine biodiversity research and surveillance.
The results of this study were presented in a poster session of the DNAqua International Conference.
The full study details can be accessed here.




Comments (0)