Marine Biosecurity Toolbox scientist Dr Ulla von Ammon joined the BLAKE Expedition and shared with the expedition team the eDNA tools, including the newly designed Cruising Speed Net, for detecting unwanted organisms in marine environments.
Right on-the-spot! How a human biologist became engaged in developing a rapid molecular test for marine pest detection
by Martin Zirngibl
I studied general biology at the Ludwig Maximilian University of Munich with the main focus on human biology and conducted my master’s thesis at the University of Alberta, Canada on the role of microglia in Multiple Sclerosis in the lab of Dr. Jason Plemel. After finishing my masters, I needed some time off and used this time to explore other places and interesting fields of research. I arrived to New Zealand in March, just before the country went into lockdown and caught the attention of the molecular biosecurity team at the Cawthron Institute in Nelson joining the Marine Biosecurity Toolbox programme as a laboratory technician in June. This gave me the opportunity to work in a new exciting field where I can contribute my knowledge and skills gained in a human biology field to help making the marine environment a nicer place, free of unwanted pests.
Busy days in the molecular lab.
Within the DETECT workstream of the Marine Biosecurity Toolbox program I am designing a rapid and easy to use detection assay for the invasive Mediterranean fanworm Sabella spallanzanii. First detected in 2008, S. spallanzanii is now present in all major shipping harbours of New Zealand and spreads mainly by hitchhiking on ship hulls. As a typical invader, S. spallanzanii is highly resistant to temperature and salinity changes. Colonies can overgrow whole ecosystems and outcompete native taxa. Therefore, detection and monitoring the spread of this species remains high on agenda of biosecurity practitioners in New Zealand.
In that context, very sensitive molecular detection techniques gain lots of interest and are being increasingly applied in New Zealand coastal waters for the fanworm detection. The current molecular detection workflow, although proven to be robust, is quite cumbersome and requires specialised laboratory facilities and skills to process the samples. Now, imagine, how cool it would be if molecular biosecurity detection approaches were simplified and adapted for the point-of-need application by end-users like regional councils, kaitiaki, schools and public volunteers that do not have lab access or enough molecular expertise, but are keen to help protect marine environment! A rapid and easy-to-apply in-field test could help engage citizen scientists and increase efficiency of the biosecurity surveillance overall.
The new detection assay I am developing is based on recombinase polymerase amplification (RPA), which uses enzymes that amplify the target DNA under isothermal conditions (constant temperature). This approach has been shown to be successful in a biosecurity framework for the Northern Pacific Seastar (Asterias amurensis) (Ravindran et al., in review).
Conventional methods of DNA amplification (i.e. real-time PCR assays) require the reaction temperature to rapidly cycle within a range of ~50 to 94 °C. This requires an expensive machine that can heat and cool the reaction as required. Specific enzymes in the RPA assay however use target specific primers (short nucleic acid sequence which initiate DNA synthesis) to find and amplify e.g. S. spallanzanii DNA at a constant (ideally 39 °C or lower) temperature. This allows end users to perform the assay while holding the reaction in the hand. The amplification is rapid (within 20 min) and yields high quantities of amplification product.
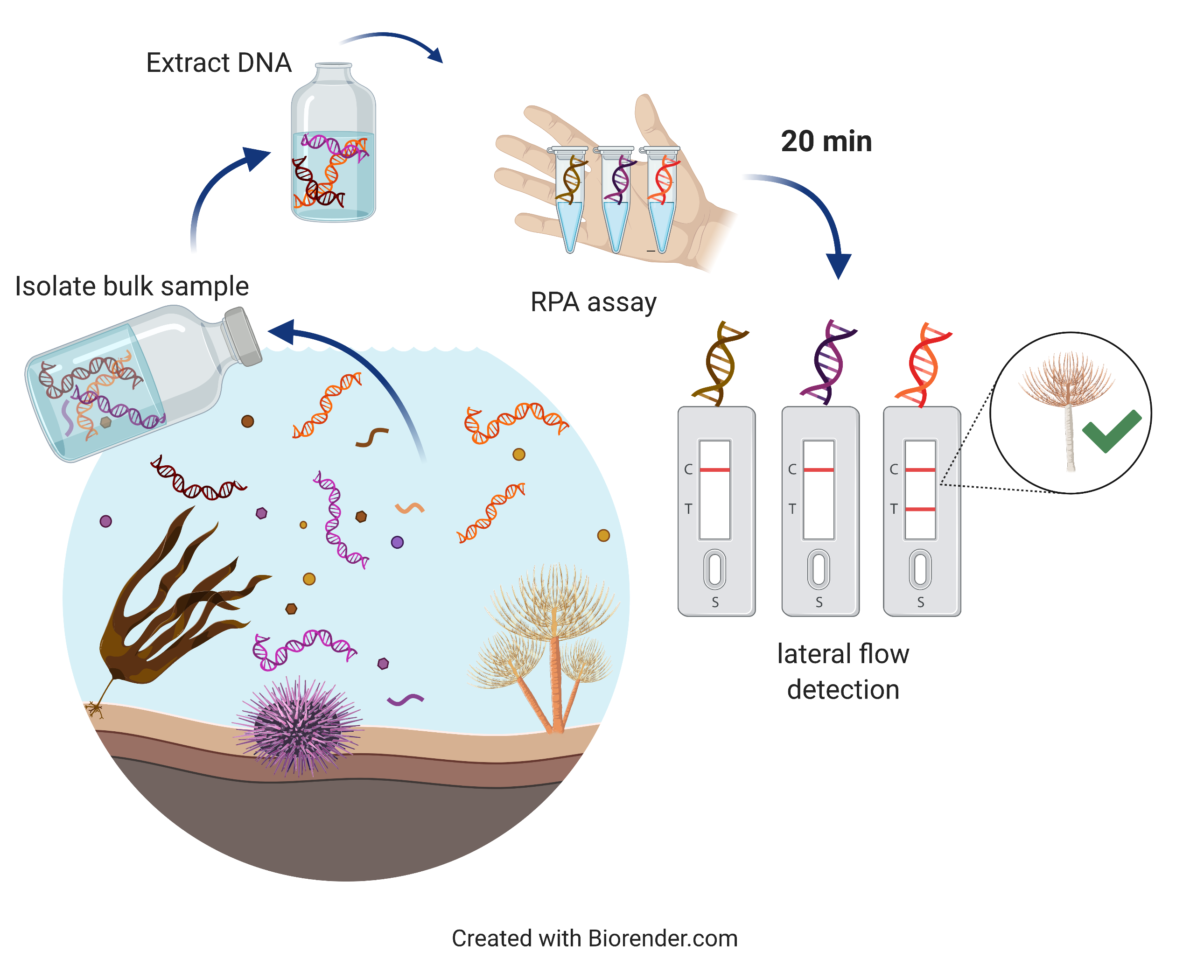
A schematic workflow of a real-time marine pest detection using an RPA assay.
I designed multiple primer combinations that are specific to S. spallanzanii and tested if they work. I used these primers in conventional PCR and RPA assays and compared both with gel electrophoresis. This technique is used to separate and visualize DNA molecules of different lengths. The PCR and RPA reaction will be applied to an agarose gel mixed with a dye that makes DNA visible under UV light. In this agarose gel DNA molecules migrate if an electric current is applied. The smaller the molecule, the further the molecule migrates in the gel. Having a size marker in the gel (blue star) you can determine the size of the DNA molecule. With this method we can determine if the primers amplify the desired region.
Below is an agarose gel showing the test of my primers with PCR (top) and RPA (bottom) on S. spallanzanii DNA. The primers I designed for the new assay are indicated by the red square, the primers that are used in the lab for detection of S. spallanzanii for ddPCR are indicated by green circle. The gel shows that the newly designed primers show the same pattern in the RPA and PCR. The ddPCR primer however show a band in the PCR, but not in the RPA. This is not surprising, because RPA primer are usually longer than ddPCR primer. This test demonstrates that the RPA primer amplify S. spallanzanii.
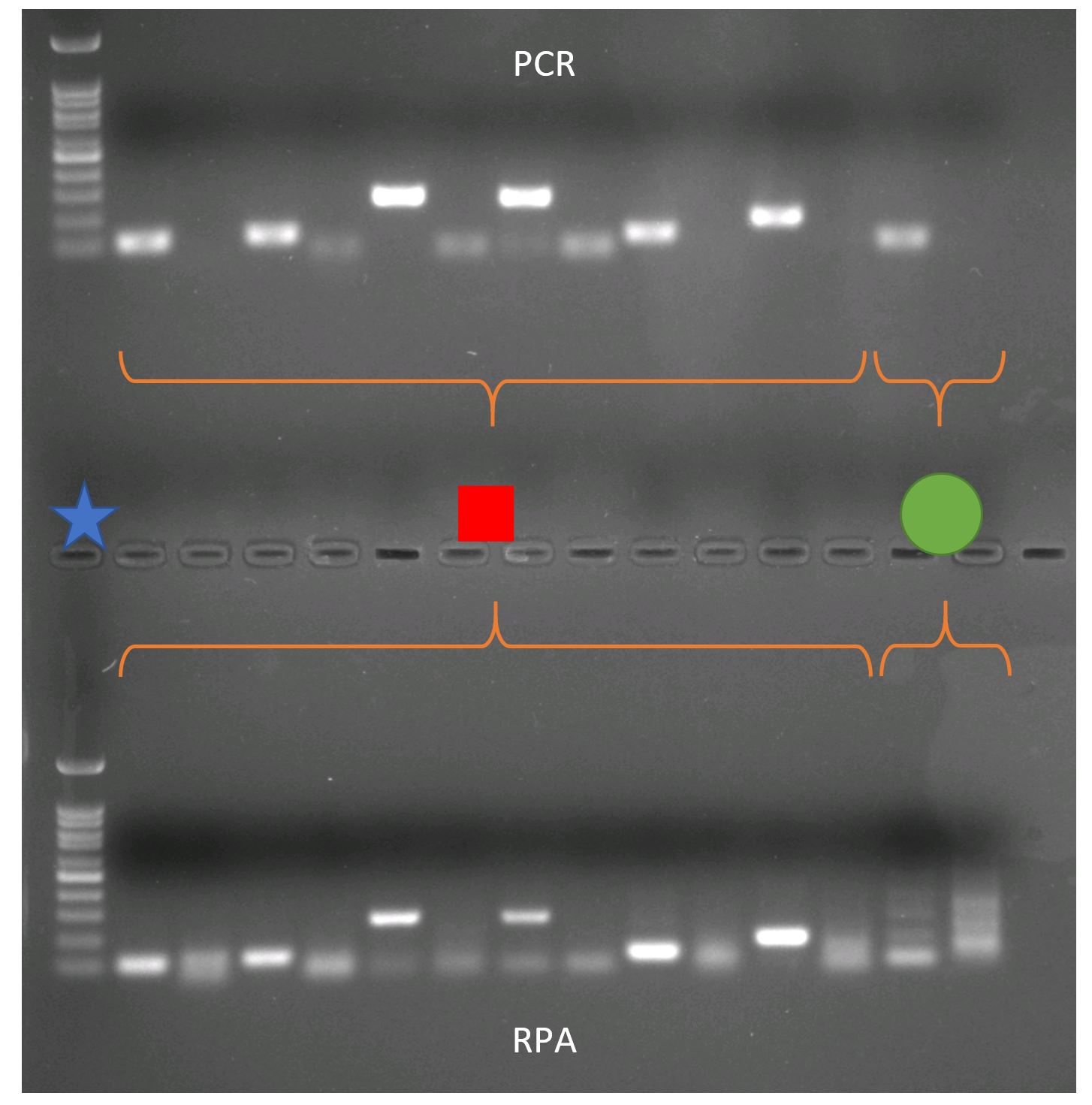
Comparison between PCR and RPA. PCR and RPA were performed using the same combinations of RPA primer and the well-established primers used for real-time PCR.
As a next step, I will test the best performing primer combinations for cross-reactivity with native New Zealand species, closely related to S. spallanzanii. This step of quality control ensures that a positive amplification can be undoubtedly be interpreted as “S. spallanzanii present in the sample.”
The final readout of the reaction will be with a conventional lateral-flow stick, which works just like a pregnancy test is then used to display positive or negative results. This fast and easy readout makes this system perfect for non-expert end-users on the ground, who do not have access to specific equipment such as gel electrophoresis chambers. Once the test-kit for S. spallanzanii proves to be successful (sensitive, specific, robust and well-performing in in-field applications), the DETECT team of the programme will work forward developing such kits for the high-priority marine pests that threaten New Zealand ecosystems.
I am happy that I have an opportunity to contribute to this research and hope that this approach will make it easier for non-scientists to participate in marine biosecurity surveillance that helps to protect precious New Zealand marine environments.




[…] with rapid, efficient and user-friendly molecular tools for marine biosecurity surveillance. Thus, Martin Zirngibl is currently leading the development of a point-of-need assay for detecting the invasive […]
[…] four specific species and three broader types of biofouling organisms. Mediterranean fan worm (Sabella spallanzanii) was mentioned by all but one of the participants, while the more generic Slime and Barnacles were […]